Training tiny models efficiently with Fast AI v2 and Pytorch
Everyone talks mostly about the next 1 billion+ parameter model, but I have lots of small, even tiny, models which still takes a while to train due to large data volumes and inefficient use of the GPU.
Efficient training
I would like to be able to train tiny or small models efficiently on my GPU. I have taken inspiration from the excellent design note available on the Pytorch website. And also the tutorial using the distributed data parallel functionality available with Pytorch.
I would like to be able to, as I usually do with Fast AI 2, simply define a model, a learner and start training:
parallel_models = 10
drm = DataParallelEnsembleModule(n=parallel_models, modelfn=RegModel)
learn = Learner(
data,
drm,
lr=lr,
cbs=[AvgWeightsCallback],
loss_func=partial(xpemloss, lossfn=MSELossFlat()),
opt_func=partial(SGD, mom=0.9),
)
learn.fit_one_cycle(4)
| epoch | train_loss | valid_loss | time |
|---|---|---|---|
| 0 | 70.470818 | 68.176102 | 00:00 |
| 1 | 63.467754 | 52.796143 | 00:00 |
| 2 | 56.377953 | 44.170441 | 00:00 |
| 3 | 51.243004 | 42.282677 | 00:00 |
Ensemble of models and weights
Training multiple models in parallel quickly leads to questions about model fit. And being efficient while training is pointless if the resulting models are not better or at least as good as the basic version. Luckily the following papers apply some form of averaging, which seems to lead to ‘better’ fitting or more general models - which typically is what we want. Or at least it shouldn’t hurt us.
Averaging Weight Leads to Wider Optima and Better Generalization this paper introduces Stochastic Weight Averaging (SWA) with the promise of better more general solutions than simple SGD. Also mentioned is the Fast Geometric Ensembling (FGE) paper which is along similar lines. The former deals with averaging of the weights while the latter introduced the cyclical learning rate.
My implementation
I would like to train as I’m used to with Fast AI, and be able to instruct the framework that I want to fit i.e. 4 models in parallel on a partitioned but otherwise similar dataset. I’ll be using synth_learner and synth_dbunch from Fast AI version 2 as my base models in this example. It makes it easy to quickly get something up and running quickly, and I’m also able to reason about the correctness without too much trouble.
Design criteria and limitations
- The base model should not need to be changed or updated - not substantially anyway. Maybe patching the code if its automatic is ok.
- The supporting functions for the loss calculation should also just take the individual loss function you would like to apply to your base model. (Maybe this is not a stringent requirement, but making it as easy as possibly to try out different loss functions is important)
- Data loading should be fairly simple, I know helper functions like RandomSplitter are super elegant. But during this first step I work under the assumption that data is already partitioned and you create individual data loaders.
The following is my version, but maybe there is a trick where you expand an extra dimension on top of your model and just collapse the parameters after the fact. Let me know if you see the obvious implementation I have overlooked.
This of course also opens inexpensive ways of experimenting with new loss functions and weight integration without having access to expensive hardware. Even if you can only fit two models in parallel on your GPU I believe it will be worth it.
As always data loading and handling becomes a major part of the implementation, and all the considerations when partitioning data found in the Pytorch tutorials applies here as well.
Related work
Mostly what I found centers around multi gpu implementations. On pytorch.org solutions are focussed on either being data parallel across multiple devices or model parallel where the model is split over multiple gpus. I have taken inspiration from the excellent design note available on the Pytorch website.
https://pytorch.org/docs/master/notes/ddp.html https://pytorch.org/tutorials/intermediate/dist_tuto.html https://pytorch.org/tutorials/beginner/blitz/data_parallel_tutorial.html
I haven’t seen or found any single gpu data parallel implementations. If you are aware of any, please get in touch. And I’ll post this on the fast ai forum to get some feedback as well. In any case it has been an instructive exercise and allowed me to dive into the libraries.
Code
The code seems to be working as is, but there are several places where I’m not completely satisfied.
First some imports
from fastai2.data.all import *
from fastai2.callback.all import *
from fastai2.torch_basics import *
from fastai2.test_utils import *
from fastai2.learner import *
from fastai2.optimizer import SGD
from torch.utils.data import *
import matplotlib.pyplot as plt
and plotting functions
def _plotfit(rmodel, ax=None, ga=None, gb=None, title="Model"):
ax.set_title(title)
# Plot all the source data, to see that it is not all the same
for src in learn.dls.dataset.sources:
ax.scatter(src.tensors[0], src.tensors[1])
# This code is very specific to our example and I just want to see
# visually how well the regression model is learned.
a = rmodel.a.detach().numpy()
b = rmodel.b.detach().numpy()
x = np.linspace(-3, 3, 20)
y = a * x + b
ax.plot(x, y, "r")
# Plot the global or averaged line
y = ga * x + gb
ax.plot(x, y, "k")
def pemplotfit(x2model):
n = int(x2model.world_size)
fig, axs = plt.subplots(n, 1, sharex=True, sharey=True, figsize=(12, n * 3))
_a, _b = [], []
for m in x2model.models():
a, b = m.a.detach().numpy(), m.b.detach().numpy()
_a.append(a)
_b.append(b)
for i, (m, ax) in enumerate(zip(x2model.models(), axs)):
_plotfit(m, title=f"M{i}", ax=ax, ga=np.average(_a), gb=np.average(_b))
Data set
Let’s create a dataset where the most important criteria is that getitem returns a tuple of tuples. Usually you return an x and y but we having parallel datasets want to return a tuple of xs and a tuple of ys. Created from independent properly partitioned data sources. This steps is fairly important, but I’ll gloss over it for now as I’m more interested in the DPEM at this point.
class DPEMSynthDataset(torch.utils.data.Dataset):
"""This is totally fake, but we try our best to make it look real.
some random data is returned which looks like a function and for this
to be realistic the lines should pretty much be similar. The main idea
is that all original data is split - by some random process - into shards
or partitions which we will fit on a model each. Therefore there will be
some similarity but they will not be completely similar.
"""
def get_data(self, n, a=2, b=3, bs=16, noise=0.1):
x = torch.randn(bs * n, 1)
return TensorDataset(x, a * x + b + noise * torch.randn(bs * n, 1))
def __init__(self, n, m=2, bs=16, offset=False, **kwargs):
"""n is the number of points in each dataset.
Note that all datasets should probably be of similar length.
m is the number of models we are trying to train.
"""
# if you want to shift the data a little do that here.
def b_fn(i):
return 3 if not offset else 3 + i
self.sources = [self.get_data(n, b=b_fn(i), bs=bs, **kwargs) for i in range(m)]
self._length = n * bs
def __getitem__(self, i):
xs, ys = [], []
for src in self.sources:
x, y = src[i]
xs.append(x)
ys.append(y)
return (tuple(xs), tuple(ys))
def __len__(self):
# All datasets should have the same length
return self._length
The Data Parallel Ensemble Module
This is where most of my uncertainty lies, as I’m trying to shortcut the functionality of a ‘normal’ module, while retaining all its functionality. The idea is simply to pass functionality on e.g. the forward pass just calls forward on each model and returns the tuple of losses.
Known limitations
The DPEM shouldn’t be used as is for inference, rather you are supposed to extract parameters and load those into your base model from where prediction should be done.
class DataParallelEnsembleModule(Module):
def __init__(self, n=2, modelfn=RegModel):
# n is the number of models we want in parallel in this ensemble.
for i in range(n):
# Pytorch needs everything as attributes to register properly
# https://pytorch.org/docs/stable/nn.html#torch.nn.Module
setattr(self, f"_pem{i}", modelfn())
self.world_size = float(n)
def models(self):
return [getattr(self, f"_pem{i}") for i in range(int(self.world_size))]
def forward(self, xs):
return tuple([m(x) for m, x in zip(self.models(), xs)])
Dealing with loss
As we now have a different loss returned - usually you would get a single tensor back - we need to enable the backward pass to work properly.
class DPEMLoss:
def __init__(self, losses):
self.losses = losses
def mean(self):
# FIXME is this merly for the Recorder?
# should I simply just do tensor(...).mean() here?
return torch.mean(torch.stack([l.mean() for l in self.losses]))
def backward(self):
# I'm guessing you can just do this: torch.autograd.backward(self.losses)
# but I just call backward for each loss for now.
# And maybe this is 'correct' as there should not be cross model dependencies here.
for l in self.losses:
l.backward()
def dpemloss(pred, *targ, lossfn=MSELossFlat()):
"""Usually the output of a loss function is a tensor with torch.autograd functionality
but here we merly record each loss and return that wrapped."""
return DPEMLoss([lossfn(p, t) for p, t in zip(pred, targ[0])])
Benefitting from the parallel modules
To get any benefit from doing this in parallel we need to somehow average the weights at some interval. At this point I’m just trying things out and would like to see some reaction on the loss later on.
This mimics, or at least should mimic, the average_gradient function found in Pytorch distributed tutorial.
class AvgWeightsCallback(Callback):
# See fastai2/callback/core.py for other options
def after_step(self):
# Parameters have just updated, and we would like to average between models.
# Sum all gradients, requires that the model is a PEM
for ps in [trainable_params(m) for m in self.model.models()]:
# Calculate average gradients, create an empty gradient object here
gradsum = torch.zeros_like(ps[0].grad.data)
for i, p in enumerate(ps):
gradsum.add_(p)
gradsum.mul_(1.0 / self.model.world_size)
# Set the averaged gradients
for p in ps:
p.grad.data.copy_(gradsum)
The learning rate and batch size etc. are totally made up, but they don’t matter much here anyway for this simple regression model.
Note that I apply an offset to each dataset and a little more noise than the synth_learner does. I want to make sure that each model fit its data.
lr = 1e-3
bs = 16
# Adjust this to have more data parallelism
parallel_models = 3
x2_valid = DPEMSynthDataset(2, m=parallel_models, bs=bs, offset=True, noise=0.3)
x2_train = DPEMSynthDataset(10, m=parallel_models, bs=bs, offset=True, noise=0.3)
data = DataLoaders.from_dsets(x2_train, x2_valid, bs=bs)
Finally lets create the DPEM model by telling it to use the RegModel as base. And create the learner with our callback and loss function.
drm = DataParallelEnsembleModule(n=parallel_models, modelfn=RegModel)
learn = Learner(
data,
drm,
lr=lr,
cbs=[AvgWeightsCallback],
loss_func=partial(dpemloss, lossfn=MSELossFlat()),
opt_func=partial(SGD, mom=0.9),
)
# Uncomment if you need to just use your cpu.
# learn.model.cpu()
learn.fit(12)
| epoch | train_loss | valid_loss | time |
|---|---|---|---|
| 0 | 21.139265 | 14.529093 | 00:00 |
| 1 | 18.302132 | 10.255535 | 00:00 |
| 2 | 15.148075 | 6.724180 | 00:00 |
| 3 | 12.290758 | 4.239456 | 00:00 |
| 4 | 9.873997 | 2.633739 | 00:00 |
| 5 | 7.900713 | 1.651942 | 00:00 |
| 6 | 6.325592 | 1.049504 | 00:00 |
| 7 | 5.073884 | 0.685499 | 00:00 |
| 8 | 4.081658 | 0.464382 | 00:00 |
| 9 | 3.294685 | 0.327957 | 00:00 |
| 10 | 2.668821 | 0.242585 | 00:00 |
| 11 | 2.169432 | 0.188930 | 00:00 |
Results
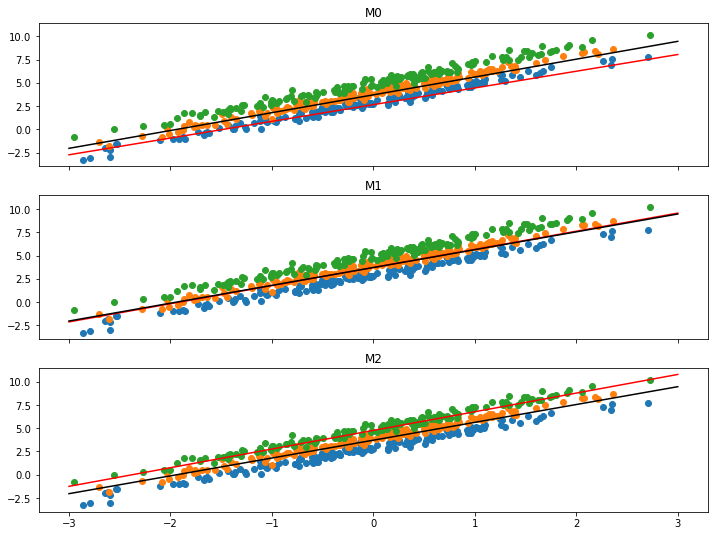
It seems to work, all models in the ensemble converge to the center of their data. I’m plotting each model with a red line showing the individual model fit as well as a black line showing the ensemble average. Without formally verifying anything, all models seem to fit fairly well - and the average prediction is centered along the middle of all the datasets - which is what I expected.
pemplotfit(learn.model)
Work in progress and next steps
This page is work-in-progress don’t expect code examples to just work.
If you have any comments please tweet me or maybe send me an email.
Todo / fixme
- The way DPEMLoss is implemented feels a little like cheating and maybe not quite right.
- Establish what mean in DPEMLoss does.
- Verify that parameter averaging in AvgWeightsCallback is correct.
- Make sure DataParallelEnsembleModule properly mimics a module. Or that I know the limitations i.e. no predict at this time.
- DPEMSynthDataset should be made more generic. (taking a function which generates one partion or something similar.)
Next steps
- Other options for when to average weights. Batch, epoch, …
- Other options for how to average weights.
- Verify that I’m actually getting the speedup I’m looking for.
- Extract a final set of parameters for inference.
- Verify this works with a more complex model. (1D convolutional network)
Last edit 09:06, 2020-06-14, Henrik Gudbrand Petersen
Changelog:
17:01, 2020-06-13: Fixed parameter copy in the callback
Appendix
Just verifying that we are changing something in the right direction. Below is shown the baseline simple regression model which I have worked from. Ideally we would like the weights to level out faster then a plain test run, as we have a larger dataset to work from.
learn2 = synth_learner()
# Running for a while should get you close to 0.01 which is the random noise which is added.
learn2.fit(12)
| epoch | train_loss | valid_loss | time |
|---|---|---|---|
| 0 | 16.111963 | 13.001087 | 00:00 |
| 1 | 14.080363 | 9.140627 | 00:00 |
| 2 | 11.805121 | 5.932382 | 00:00 |
| 3 | 9.690201 | 3.650169 | 00:00 |
| 4 | 7.855214 | 2.194892 | 00:00 |
| 5 | 6.331043 | 1.299153 | 00:00 |
| 6 | 5.091357 | 0.759722 | 00:00 |
| 7 | 4.092839 | 0.439133 | 00:00 |
| 8 | 3.292417 | 0.252125 | 00:00 |
| 9 | 2.651540 | 0.145483 | 00:00 |
| 10 | 2.138982 | 0.083558 | 00:00 |
| 11 | 1.728382 | 0.049345 | 00:00 |
We often see loss here settle after 6-10 epochs depending on the random initialization.